MM-HealthFair: Understanding Fairness and Explainability in Multimodal Approaches within Healthcare
Innovations in digital health technologies continue to drive novel applications of Multimodal Artificial Intelligence (MMAI). These approaches showcase impressive prognostic performance for risk assessment, leading to improved opportunities for delivering healthcare. However, despite their superior discrimination ability over unimodal algorithms, MMAI approaches risk inheriting and amplifying hidden biases within routine healthcare data. Increasing understanding of fairness in model decisions is a crucial part of ensuring the equitable and safe use of AI-driven risk assessments in clinical practice. This project aims to explore fairness and explainability in the context of multimodal AI for healthcare (see project description here).
The MM-HealthFair framework was developed to provide a clear evaluation of multimodal fairness and explainability, focusing on understanding driving factors behind MMAI decisions in healthcare. It leverages the MIMIC-IV open database for risk prediction in urgent care, supporting use of tabular, time-series and free-text data. The fairness toolkit provides a statistically-grounded validation of established fairness metrics for risk prediction, and an in-model adversarial mitigation mechanism to correct for biases in underrepresented groups. It additionally provides aggregates of Shapley Additive eXplanations (SHAP) estimates for interpreting decisions in a multimodal scenario and determining relative modality dependence.
MM-HealthFair is applicable for examining healthcare disparities and detecting patterns of attribution bias within sensitive groups. Fairness constraints can be enforced to mitigate the risk of introducing and amplifying these disparities within the training procedure of an MMAI algorithm. The output of this work aims to promote the dissemination of knowledge regarding fairness in MMAI algorithms, working towards ensuring transparent and equitable AI decisions. The framework contains the following key components: - Feature extraction and preparation for multimodal risk prediction using tabular (EHR), time-series and free-text data. - Multimodal algorithm development using intermediate fusion (feature concatenation). - Performance and Risk stratification analytics for binary classification. - Fairness analytics with bootstrapping. - Deep adversarial mitigation for inserting multimodal fairness constraints. - Explainable AI (SHAP) analytics for multimodal feature importance and aggregated measures for modality dependence (MM-SHAP).
For a detailed walkthrough of the framework, please refer to the Getting Started page.

Data curation
This toolkit provides a reproducible feature extraction pipeline for the fusion of three data modalities: tabular health records, time-series and free-text data. It uses the MIMIC-IV dataset (version 3.1) to define prediction objectives for risk classification in hospitalised patients at point of arrival to the emergency department (ED). These include prediction of in-hospital death, extended stay, non-home discharge and admission to ICU. Target sensitive attributes to explore during the fairness analysis and debiasing process can be set between gender, ethnicity, marital status and insurance. The data curation and preprocessing is performed using the following scripts:
extract_data.py: Reads and filters relevant hospital stay data from downloaded MIMIC-IV files.prepare_data.py: Cleans and prepares features in unique patients for multimodal learning into a .pkl files for downstream analysis. This will also generate a training, validation and testing set for the specified risk prediction task.
Configurations regarding the target objective, the sensitive attributes and any additional metadata for visualisation can be customised by editing the targets.toml file.
Multimodal learning and validation
In this project, we use an intermediate fusion approach to fuse the three data modalities using concatenation with equal weight. Each modality is trained on a separate deep neural net component and fused at the final hidden layer. The currently supported network components are: - MM-EHR (MLP): Multi-layer Perceptron classifier for tabular data. - MM-TS (LSTM): 2-component LSTM classifier (for vital signs and lab measurements data). - MM-NT (TF-E): Transformer-encoder network for free-text (discharge summaries) data.
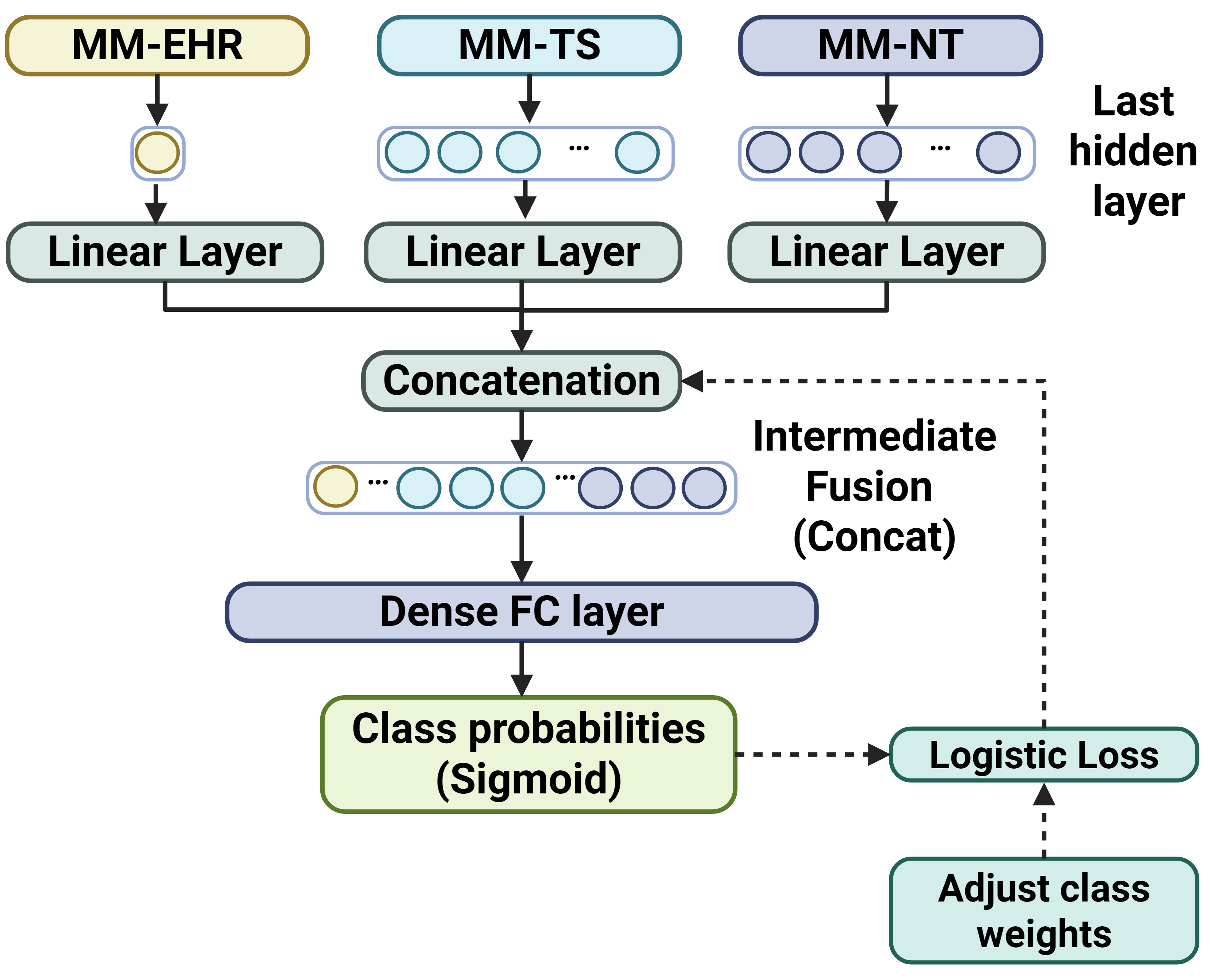
After training a multimodal algorithm, we can execute the evaluation pipeline for performance assessment. To run the pipeline we use:
train.py: Script to train a deep neural net for risk prediction. Option to log and save models using Weights & Biases.evaluate.py: Run model inference including performance/calibration/loss summaries with confidence intervals, including risk stratification to set risk quantiles per patient in the test set.
Training configurations are specified in a config file. See model.toml for available settings. Fusion method can set as: None (allows unimodal learning), concat (feature concatenation) or mag (multi-adaptation gates - works only when fusing tabular and time-series data). To enable adversarial mitigation, we set the adv_lambda above 0 within model.toml, then specify the target sensitive attribute ids in targets.toml (dbindices) add finally use the --use_debiasing argument when running train.py. This will include additional adversarial head units per attribute, reducing their influence within the tabular modality.
Fairness and Explainability analytics
Once models have been trained and saved, we also include the scripts used to quantify fairness and examine explainability using local and global multimodal feature importance interfaces. The Fairlearn package is used for fairness evaluation, while the SHAP package is used to compute and aggregate Shapley values as multimodal feature importance scores. The supported fairness metrics include demographic parity (DPR), equalised odds (EQO) and equal opportunity (EOP). These can be estimated using confidence intervals with customisable bootstrapping iterations via the BCa method. Global-level explanation plots can be generated via SHAP density or heatmap plots for tabular and time-series or risk barchart plots highlighting important note segments. On the local-level, we can set a target patient profile to randomly sample, with sensitive attributes set in targets.toml and the target level of observed risk (estimated using evaluate.py). The script will attempt to extract the SHAP values for a patient matching this profile and visualise decision and note segment plots highlighting degree of modality dependence (MM-SHAP).
fairness.py: Run the fairness inference and store results with bootstrapped samples in a binary.pkldictionary.explain.py: Run the explainability inference, with--exp_modein eitherglobalorlocalmode and save the multimodal feature importance plots.