Important: Disclaimer
This is not the official site but a set of brief descriptions of our recent work to support transparency and collaboration. For more information about NHS England please visit our official website
Investigating Superpixels in LIME for Explaining Predictions of Facial Images
“Investigating explainability algorithms for granular healthcare images”
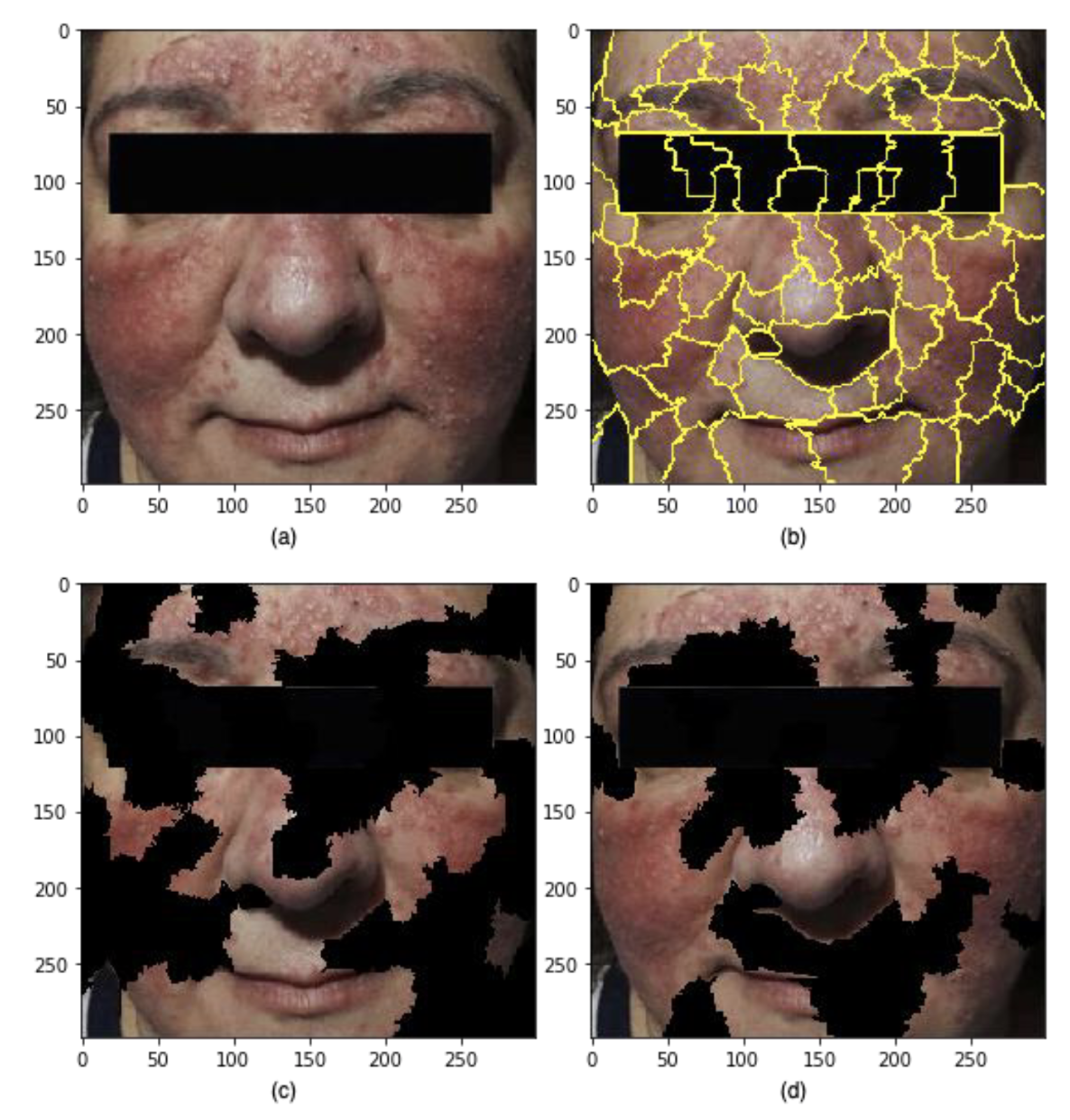
Figure 1: Output from the LIME pipeline for a Rosacea face. 3(a) is an example image from the rff-300 dataset; 3(b) is an illustration of 73 superpixels found in the image; 3(c) is an illustration of random perturbations of the superpixels; 3(d) show the top features used in training to created the best performing predictions.
A work experience project investigating the application of a Local Interpretable Model-agnostic Explanations (LIME) technique to an image classification task around identifying Rosacea. A binary classification model was trained on the normal and Rosacea faces to generate the LIME explanation for Rosacea faces. Secondly, the fine-tuned model was integrated into the LIME pipeline to generate explanations based on the crucial features on which predictions were made in the classification model. Hence the experimentations helped in understanding the features the classification model took.
Results
Image pre-processing (through contrast enhancement) improved the LIME by increasing the number of superpixels for the given input image so that some broad features usch as discolouration and rashes were identified. However, the coarseness of the superpixels generated through the pipeline was not sufficient to pick up the features which would disceren between different types of Rosacea. It may require domain/ imaging modality-specific pre-processing tasks to enhance the quality of explanation by improving the distinctiveness of the features that may help pick the right number of superpixels.
| Output | Link |
|---|---|
| Open Source Code & Documentation | Github |
| Case Study | n/a |
| Technical report | Here |
 |
 |